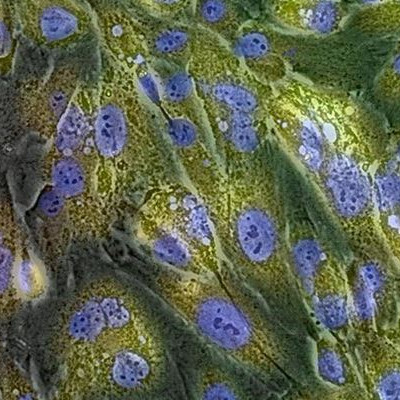
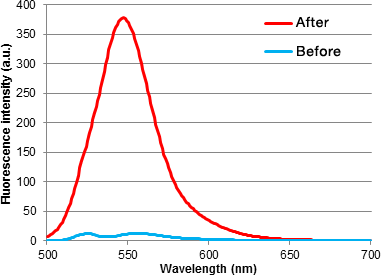
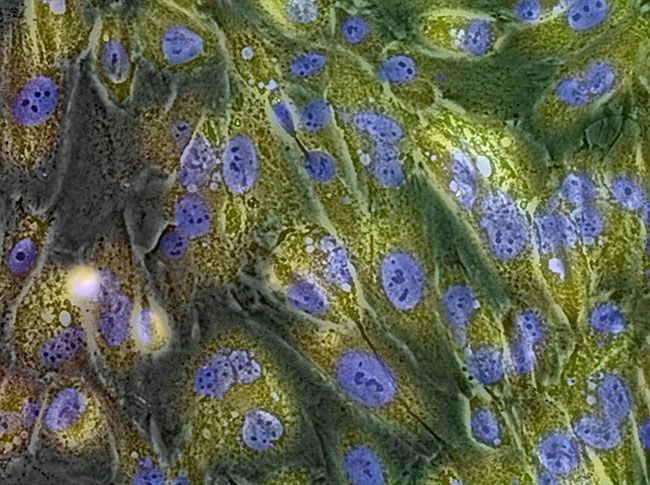
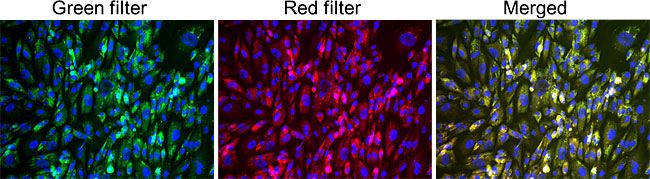
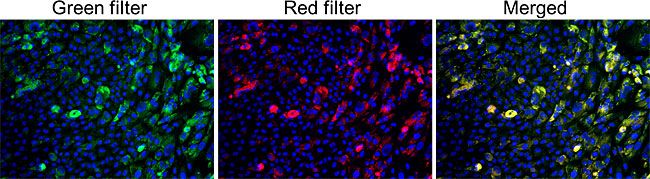
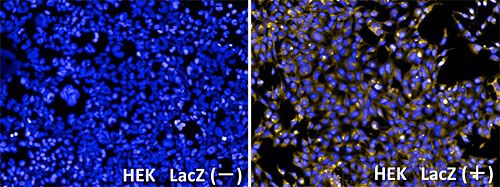
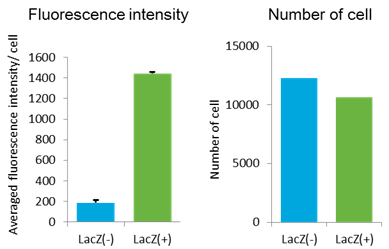
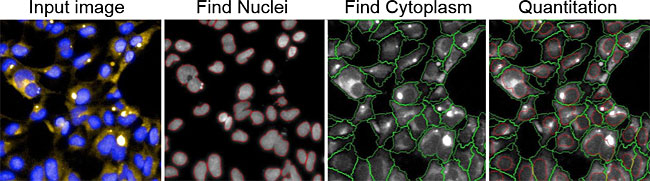
GlycoYELLOW-βGal is a fluorescent ligand for β-galactosidase. It shows almost no fluorescence but fluoresces upon the reaction with β-galactosidase. It can be applied to fluorescence imaging and selection of lacZ-gene expressing cells and tissues. In addition, it can be used to detect senescence-associated β-galactosidase (SA-β-Gal) activities.
Available through Merck KGaA (Darmstadt, Germany ) as:
SCT024 BioTracker™ 543 Yellow β-Gal Dye
![]()





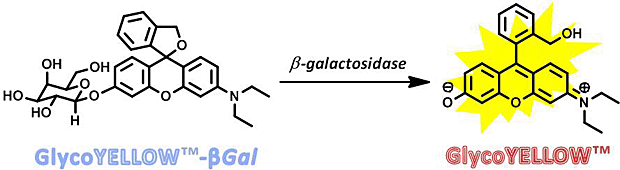
 Contact Us
Contact Us
