R. Furuuchi, I. Shimizu, Y. Yoshida, Y. Hayashi, R. Ikegami, M. Suda, G. Katsuumi, T. Wakasugi, M. Nakao, T. Minamino (2018)
PLOS one 13:e0202051 DOI: 10.1371/journal.pone.0202051
M. Kobayashi, M. Miyamoto, T. Matoh, S. Kitajima, S. Hanano, I.N. Sumerta, T. Narise, H. Suzuki, N. Sakurai, D. Shibata(2018)
J Soil Sci Plant Nut. 64: 1, 106-115 DOI: 10.1080/00380768.2017.1416670
J. Anstine, C. Bobba, S. Ghadiali, J. Lincoln (2016)
Mol. Cell Cardiol. 100: 72–82. DOI:10.1016/j.yjmcc.2016.10.006.
N. Yousfi , B. Pruvot, T. Lopez, L. Magadoux, N. Franche, L Pichon, F. Salvadori, E. Solary, C. Garrido, V. Laurens, J. Chluba (2015)
PLoS One. 10:e0120435. DOI: 10.1371/journal.pone.0120435
N. Gan, T. Hondou, H. Miyata (2012)
Biol. Pharm. Bull. 35: 1454-1459 DOI:10.1248/bpb.b11-00010
F. J. Corpas, M. Hayashi, S. Mano, M. Nishimura, J. B. Barroso (2009)
Plant Physiol. 151: 2083-2094 DOI:10.1104/pp.109.146100
T. Nagano (2009)
J. Clin. Biochem. Nutr. 45: 111-124 DOI:10.3164/jcbn.R09-66
J. R. Steinert, C. Kopp-Scheinpflug, C. Baker, R. A. Challiss, R. Mistry, M. D. Haustein, S. J. Griffin, H. Tong, B. P. Graham, I. D. Forsythe (2008)
Neuron 60: 642-656 DOI:10.1016/j.neuron.2008.08.025
A. Vatsa, R. G. Breuls, C. M. Semeins, P. L. Salmon, T. H. Smit, J. Klein-Nulend (2008)
Bone 43: 452-458 DOI:10.1016/j.bone.2008.01.030
X. Ye, S. S. Rubakhin, J. V. Sweedler (2008)
Analyst 133:423-433 DOI:10.1039/b716174c
S. Lepiller, V. Laurens, A. Bouchot, P. Herbomel, E. Solary, J. Chluba (2007)
Free Radic. Biol. Med. 43: 619-627 DOI:10.1016/j.freeradbiomed.2007.05.025
A. Vatsa, D. Mizuno, T. H. Smit, C. F. Schmidt, F. C. MacKintosh, J. Klein-Nulend (2006)
J. Bone Miner. Res. 21: 1722-1728 DOI:10.1359/jbmr.060720
N. N. Tun, C. Santa-Catarina, T. Begum, V. Silveira, W. Handro, E. L. FlohI, G. F. Scherer (2006)
Plant Cell Physiol. 47: 346-354 DOI:10.1093/pcp/pci252
C. Vecchione, A. Aretini, G. Marino, U. Bettarini, R. Poulet, A. Maffei, M. Sbroggiò, L. Pastore, M. T. Gentile, A. Notte, L. Iorio, E. Hirsch, G. Tarone, G. Lembo (2006)
Circ. Res. 98: 218-225 DOI:10.1161/01.RES.0000200440.18768.30
C. Núñez, V. M. Víctor, R. Tur, A. Alvarez-Barrientos, S. Moncada, J. V. Esplugues, P. D’Ocón (2005)
Circ. Res. 97: 1063-1069 DOI:10.1161/01.RES.0000190588.84680.34
M. Satoh, S. Fujimoto, Y. Haruna, S. Arakawa, H. Horike, N. Komai, T. Sasaki, K. Tsujioka, H. Makino, N. Kashihara (2005)
Am. J. Physiol. Renal. Physiol. 288: F1144- F1152 DOI:10.1152/ajprenal.00221.2004
H. Kojima, M. Hirotani, N. Nakatsubo, K. Kikuchi, Y. Urano, T. Higuchi, Y. Hirata, Nagano, T (2001)
Anal. Chem. 73: 1967-1973 DOI:10.1021/ac001136i
R. Furuuchi, I. Shimizu, Y. Yoshida, Y. Hayashi, R. Ikegami, M. Suda, G. Katsuumi, T. Wakasugi, M. Nakao, T. Minamino (2018)
PLoS One 13: e0202051 DOI:10.1371/journal.pone.0202051


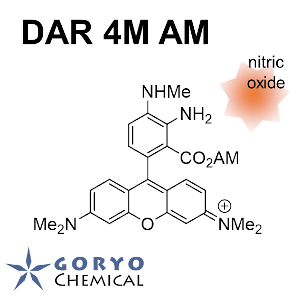



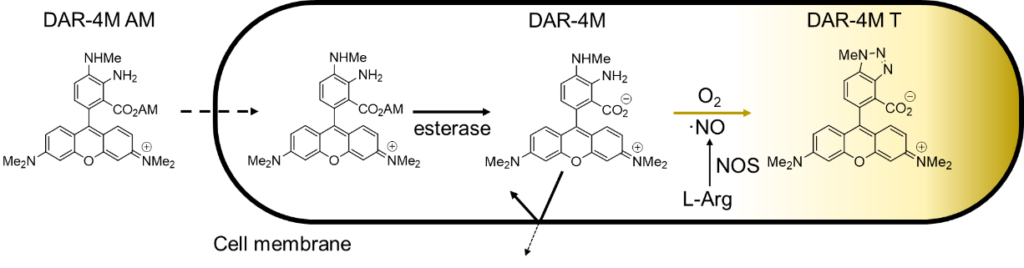
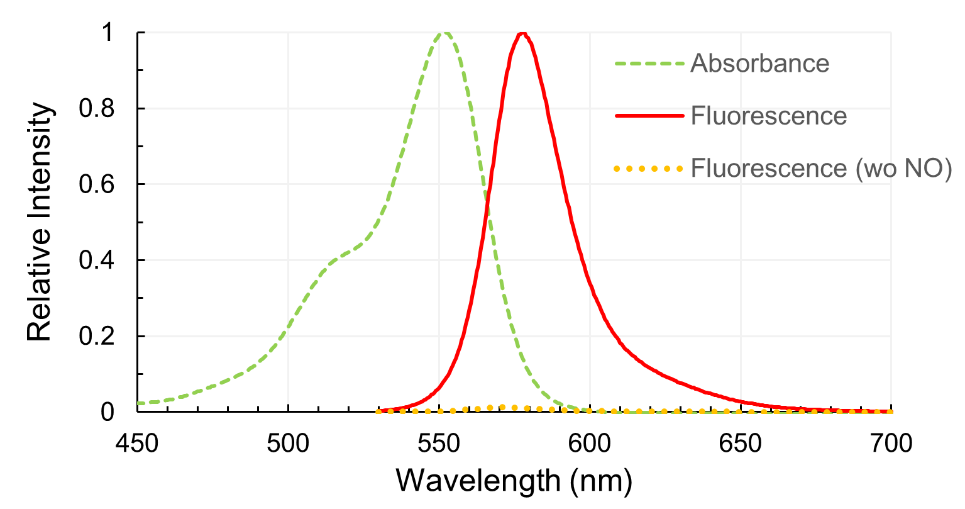
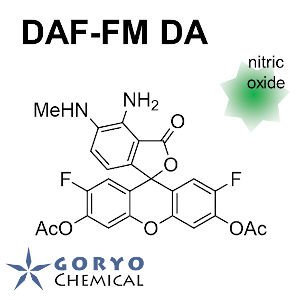
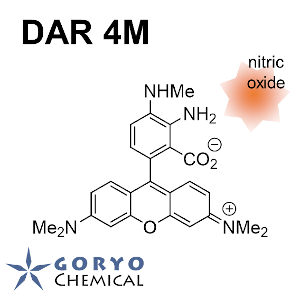
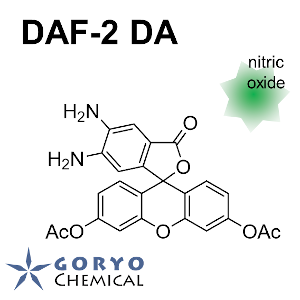
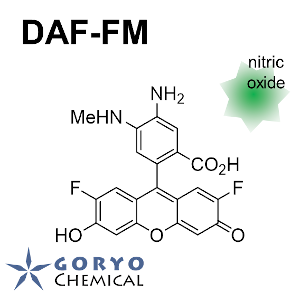
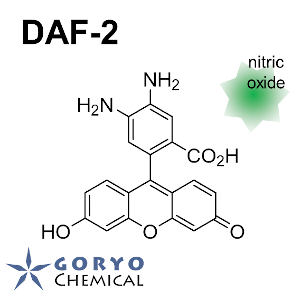
 Contact Us
Contact Us
